Pipeline¶
Welcome to the PLATO Ecosystem: A Docker wrapper for PlatoSim3 and the LESIA L1 pipeline. The following guide explains how to install, setup, and run the LESIA L1 pipeline as an integrated part of PLATOnium. Courtesy goes to Réza Samadi (LESIA), who led the code development for the pipeline, and to James McCormac (UW), who developed the Docker Ecosystem.
Initial setup¶
- Install docker and docker-compose following the instructions for your host OS.
- Clone the GitHub docker ecosystem repository to your server
- Inside the
docker_ecosystemfolder clone thePlatoSim3repo (checkout whatever branch you currently want to use, e.g. develop) - Inside the
docker_ecosystemfolder create a folder calledalgos - Inside the
algosfolder clone the LESIA L1 pipelinecommonrepository - Speak to KUL developers, LESIA developers, and James McCormac for access if required to PlatoSim3, L1 pipeline, and Ecosystem, respectively.
The software packages are cloned into this parent level directory to avoid passing git credentials into the Docker images. Once cloned the codes are copied into the Docker image as part of the setup. See image below for a schematic of the system.
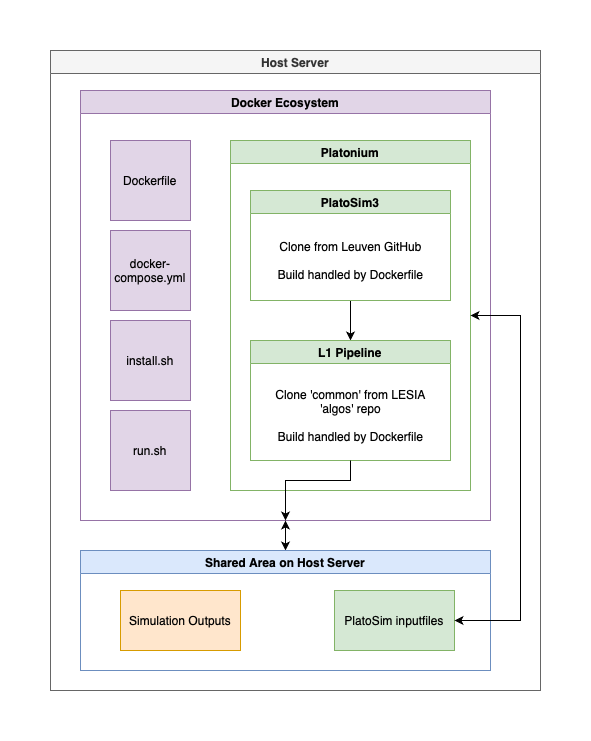
Installation¶
Building the Docker image¶
- From the
docker_ecosystemparent level folder do the following - Run
./install.sh - An Ubuntu 20.04 image will be created with the following actions:
- Python 3.9
- Sets up a non-root user etc
- Linux libraries for PlatoSim3 and L1 the pipeline
- Python modules for PlatoSim3 and L1 the pipeline
- PlatoSim3 and the L1 pipeline themselves
- Building an image from scratch with no caching takes about 30 min.
Configuring simulation storage area on host¶
Edit the docker-compose.yml file, adding a path on your host machine where you’d like to save the data. Specifically, edit the line in the volumes: section to map /path/on/host/:/host_dir. Docker will mount this area when the container stars and results will persist when the container is stopped.
Starting or stopping a container¶
Execute the
run.shscript to spin up a container in interactive mode- This uses
docker-composeto mount the storage area inside a container - Simulations can then be run inside the container as normal (see below)
- This uses
Type
exitto quit the container (as if it was a normal terminal)
Updating software in the Docker image¶
If one of the three software packages is updated, simply pull the latest code into the ecosystem folder and rerun the ./install.sh
command.
Pruning Docker resources¶
If you use Docker to build many images it’s advisable to run:
docker system prune -a
occasionally to free up some resources. Docker caches image layers to increase build speed but after a while those layers become stale as images are updated.
Run example¶
In this example we show hot to run the PlatoSim + L1 Pipeline in the container. Platonium is a wrapper around both PlatoSim and the L1 pipeline. Below is the current usage for platonium and an example command to simulate 1 quarter of data for a given star on a particular camera including some supplied variability.
Platonium requires the normal PlatoSim inputfile.yaml along with a catalog from picsim, any variability signals (e.g. from varsim) and instrument specific configurations for multiple cameras from payload. Please see the full platonium documentation for a full description of the inputfiles. Note, picsim, varsim and payload can be run inside the container environment also. Platonium expects these input files in the following directory:
/host_dir/<project>/input
where <project> is described below and /host_dir is the folder mounted from the host OS in the docker-compose.yml file above.
The following command can be run inside the container to produce a quarter long simulation of a single star (number 46) from camera group 1 camera 1 from quarter 23 and assuming the P5 sample. We also inject stellar/transit signals using the --varfile. The project kul20 corresponds to the KUL technical note 20 simulation settings, therefore for this run the inputs should be stored in /host_dir/kul20/input.
platonium 46 1 1 23 --project kul20 --sample P5 --pipeline --varfile /host_dir/varsource/P5/varsource_000000046.txt -v 3 -w
Platonium runs PlatoSim and analyses the imagettes. Outputs (photometry etc) are stored in /host_dir/kul20/output/reduced/P5/000000046/. Platonium also produces a summary of the processing time when complete.
Output files¶
During a simulation, PLATOnium creates
three folders called reduced,
microscan,
P1
(or P5
depending on sample parsed). By default during the run a lot of
files are created and stored in microscan
and P1,
but at end simulation, the final files are stored into reduced.
Note that these foder contains a tree of subfolders to keep each
simulation isolated when running in parallel. All data products are
saved using the HDF5 format. A standard filename is used for each
data product, e.g. the light curve file:
<PLATOID>_Ncam<camera>_Q<quarter>_LIGHTCURVE_L1A_IMAGETTE.hdf5
(for P1 sample)
<PLATOID>_Ncam<camera>_Q<quarter>_LIGHTCURVE_L1A.hdf5 (for P5 sample).
For a general overview of the on-board and on-ground pipeline see PLATO Synopsis of on-board processing algorithms (PLATO-LESIA-PDC-RP-0024, i2.8) and PLATO Synopsis of on-ground processing algorithms (PLATO-LESIA-PDC-RP-0023, i1.6) respectively.
Data product name |
Description |
Main documents |
|
Individual L1 lightcurve generated with the PSF-fitting method from imagette time-series. |
PLATO-IRAP-PDC-DD-0056 (i1.3) and PLATO-IRAP-PDC-TN-0056 (i1.2) |
|
Individual lightcurve generated on-board with aperture mask and corrected on-ground by the L1 pipeline |
Marchiori et al (2019), PLATO-LESIA-PDC-DD-0008 (i2.5), PLATO-USP-PDC-DD-0001 (i1.0) and PLATO-LESIA-PDC-DD-0043 (i1.3) |
COB_OG |
Center Of Brightness (COB) time-series generated (on-ground) with the PSF-fitting method. |
PLATO-IRAP-PDC-DD-0056 (i1.3) and PLATO-IRAP-PDC-TN-0056 (i1.2) |
COB_L0 |
Center Of Brightness (COB) time-series generated on-board with aperture mask.
|
PLATO-LESIA-PDC-DD-0008 (i2.5) |
SKYPOS_L1A |
Sky positions time-series inferred from the star COB time-series. |
PLATO-UOL-PDC-DD-0007 (i1.9) and PLATO-LIRA-PDC-TN-0095 (i1.0) |
inverse_psf |
Inverse PSF
|
PLATO-LESIA-PDC-RP-0036 (i1.2), PLATO-LESIA-PDC-DD-0039 (i1.2), PLATO-LESIA-PDC-DD-0053 (i1.1) |
interpolated_psf |
Interpolated PSF
|
PLATO-LIRA-PDC-DD-0071 (i1.0) |
target_star |
Information about the target star and its neighborhood stars |
|
Ligthcurve data product
Aperture-based lightcurve
Metadata
These metadata are used for light curves produced using aperture mask photometry i.e. produced on-board or for moderately saturated stars.
|
Description |
Type |
Unit |
WINDOW_ID |
Id of the window |
uint32 |
[-] |
PLATOID |
PLATO ID of the target |
uint64 |
[-] |
CAMERA_ID |
ID of the camera (1 to 26) |
uint8 |
[-] |
CCD_ID |
ID of the CCD
|
uint8 |
[-] |
CCD_SIDE |
Side of the CCD |
uint8 |
[-] |
SAMPLING |
Sampling of the flux time-series (typical values 25s, 50s, 600s) |
double |
second |
FLUX_ORIGIN |
Bit array giving the origin of this flux |
uint8 |
[-] |
F_PROCESSING |
Bit array giving the processing undergone by the flux |
uint16 |
[-] |
I_PROCESSING |
Bit array giving the processing undergone by the imagette Relevant if this flux comes from aperture photometry on on-ground processed imagette |
uint16 |
[-] |
TARGET_STAR_MASK_TS |
Information about the MASKs (nominal and extended) of a target as a function of time |
TARGET_STAR_MASK_TS Struct |
[-] |
Data contain
Description of a flux of a target at a given time
|
Description |
Type |
Unit |
BARYCENTRIC_TIME |
Timestamp (barycentric) when the flux (or the imagette if on-ground imagette photometry) has been acquired onboard. Expressed in Julian days since J2000_TIME_ORIGIN. |
double |
Julian day |
ONBOARD_TIME |
Timestamp based on the on-board time when the flux (or the imagette if on-ground imagette photometry) has been acquired onboard. This is the time of the beginning of the readout of the first exposure (if several exposure have been used), expressed in seconds since ON_BOARD_TIME=0. |
double |
second |
FLUX |
Value of the flux for this exposure time |
double |
|
FLUX_VARIANCE |
Value of the variance of the flux for @LongCadence exposures |
double |
ADU²
before gain correction |
CHI2 |
Chi2 of the flux, generated by the PSF fitting algorithm |
double |
[-] |
EXPOSURE_ERROR_ARRAY |
Bit mask of exposure in error for this flux. Typically, the exposure error flag is encoded on 2 bits for short cadence (2 exposures) and on 24 bits for long cadence (24 exposures) to encode which of the 2 or 24 exposures have been discarded during on-board averaging. Bit value is True (i.e. 1) if an outlier has been detected on the exposure. Only relevant when using aperture mask photometry |
uint32 |
[-] |
BACKGROUND |
Level of background for this flux |
double |
e-/exposure |
SPR_TOT |
Stellar Pollution Ratio total as defined in Marchiori et al (2019) Only relevant when using aperture mask photometry |
double |
[-] |
LTD_JIT_COR_MEAN_FACTOR |
Long-term drift and jitter noise correction factor Only relevant when using aperture mask photometry |
double |
[-] |
IMAGETTE_OUTLIERS_NB |
Number of pixels flagged as outliers in imagette. Only relevant for flux coming from photometry of imagette. |
uint16 |
[-] |
STATUS |
Status of the measurement (See STATUS Definition) |
uint32 |
[-] |
PSF-based lightcurve data product
Metadata
These metadata are used for light curves produced on-ground, i.e: using PSF photometry on imagette
|
Description |
Type |
Unit |
WINDOW_ID |
Id of the window |
uint32 |
[-] |
PLATOID |
PLATO ID of the target |
uint64 |
[-] |
CAMERA_ID |
ID of the camera (1 to 26) |
uint8 |
[-] |
CCD_ID |
ID of the CCD
|
uint8 |
[-] |
CCD_SIDE |
Side of the CCD |
uint8 |
[-] |
SAMPLING |
Sampling of the flux time-series (typical values 25s, 50s, 600s) |
double |
second |
CONTAMINANTS_LIST |
List of instance of the CONTAMINANTPFLUX structure. Each instance gives some information related to a given neighborhood star. Empty list if no contaminant stars. |
CONTAMINANTPFLUX struct
(Nc) |
[-] |
FLUX_ORIGIN |
Bit array giving the origin of this flux |
uint8 |
[-] |
F_PROCESSING |
Bit array giving the processing undergone by the flux |
uint16 |
[-] |
I_PROCESSING |
Bit array giving the processing undergone by the imagette |
uint16 |
[-] |
Data contain
Description of a flux of a target at a given time
|
Description |
Type |
Unit |
BARYCENTRIC_TIME |
Timestamp (barycentric) when the flux (or the imagette if on-ground imagette photometry) has been acquired onboard. Expressed in Julian days since J2000_TIME_ORIGIN. |
double |
Julian day |
ONBOARD_TIME |
Timestamp based on the on-board time when the flux (or the imagette if on-ground imagette photometry) has been acquired onboard. This is the time of the beginning of the readout of the first exposure (if several exposure have been used), expressed in seconds since ON_BOARD_TIME=0. |
double |
second |
FLUX |
Value of the flux for this exposure time |
double |
|
FLUX_VARIANCE |
Value of the variance of the flux for @LongCadence exposures |
double |
ADU²
before gain correction |
CHI2 |
Chi2 of the flux, generated by the PSF fitting algorithm |
double |
[-] |
EXPOSURE_ERROR_ARRAY |
Bit mask of exposure in error for this flux. Typically, the exposure error flag is encoded on 2 bits for short cadence (2 exposures) and on 24 bits for long cadence (24 exposures) to encode which of the 2 or 24 exposures have been discarded during on-board averaging. Bit value is True (i.e. 1) if an outlier has been detected on the exposure. Only relevant when using aperture mask photometry |
uint32 |
[-] |
BACKGROUND |
Level of background for this flux |
double |
e-/exposure |
SPR_TOT |
Stellar Pollution Ratio total as defined in Marchiori et al (2019) Only relevant when using aperture mask photometry |
double |
[-] |
LTD_JIT_COR_MEAN_FACTOR |
Long-term drift and jitter noise correction factor Only relevant when using aperture mask photometry |
double |
[-] |
IMAGETTE_OUTLIERS_NB |
Number of pixels flagged as outliers in imagette. Only relevant for flux coming from photometry of imagette. |
uint16 |
[-] |
STATUS |
Status of the measurement (See STATUS Definition) |
uint32 |
[-] |
Contaminant lightcurve
|
Description |
Type |
Unit |
FREE_AMPLITUDE |
Boolean indicating if the contaminant amplitude is a free parameter of the PSF fitting model |
bool |
[-] |
FLUX_TS |
Flux time series of the contaminant |
FLUX Struct |
[-] |
Center Of Brightness (COB) data product
Metada
Centre of Brightness of a single target metadata
|
Description |
Type |
Unit |
WINDOW_ID |
Id of the window |
uint32 |
[-] |
PLATOID |
PLATO ID of the target |
uint64 |
[-] |
CAMERA_ID |
ID of the camera (1 to 26) |
uint8 |
[-] |
CCD_ID |
ID of the CCD
|
uint8 |
[-] |
CCD_SIDE |
Side of the CCD |
uint8 |
[-] |
SAMPLING |
Sampling of the COB time-series (typical value 25s, 50s, 600s) |
double |
second |
CONTAMINANTS_LIST |
List of instance of the CONTAMINANTPCOB structure. Each instance gives some information related to a given neighborhood star. Empty list if no contaminant stars. |
CONTAMINANTPCOB struct
(Nc) |
[-] |
COB_ORIGIN |
Bit array giving the origin of this COB |
uint8 |
[-] |
C_PROCESSING |
Bit array giving the processing undergone by the COB |
uint16 |
[-] |
I_PROCESSING |
Bit array giving the processing undergone by the imagette, relevant if this COB comes from aperture photometry on on-ground processed imagette |
uint16 |
[-] |
Data contain
Centre of Brightness of a single target for an exposure time
|
Description |
Type |
Unit |
BARYCENTRIC_TIME |
Timestamp (barycentric) when the COB (or measurements to derive the COB) has been acquired onboard. Expressed in Julian days since J2000_TIME_ORIGIN (i.e. ONBOARD_TIME=0). |
double |
Julian day |
ONBOARD_TIME |
Timestamp based on the on-board time when the COB (or measurements to derive the COB) has been acquired onboard. This is the time of the beginning of the readout of the first exposure (if several exposure have been used), expressed in seconds since ON_BOARD_TIME=0. |
double |
second |
COB_X |
COB X coordinates on the CCD in the CCD-RF |
double |
pixels |
COB_Y |
COB Y coordinates on the CCD in the CCD-RF |
double |
pixels |
COB_VARIANCE_X |
Value of the variance of the COB along X axis for @LongCadence exposures |
double |
pixels² |
COB_VARIANCE_Y |
Value of the variance of the COB along Y axis for @LongCadence exposures |
double |
pixels² |
EXPOSURE_ERROR_ARRAY |
Bit mask of exposure in error for this COB. Typically, the exposure error flag is encoded on 2 bits for short cadence (2 exposures) and on 24 bits for long cadence (24 exposures) to encode which of the 2 or 24 exposures have been discarded during on-board averaging. Bit value is True (i.e. 1) if an outlier has been detected on the exposure. |
uint32 |
[-] |
BACKGROUND |
Level of background for this COB |
double |
e-/exposure |
IMAGETTE_OUTLIERS_NB |
Number of pixels flagged as outliers in imagette. Only relevant for COB coming from photometry of imagette. |
uint16 |
[-] |
STATUS |
Status of the measurement (See STATUS Definition) |
uint32 |
[-] |
Contaminant COB
|
Description |
Type |
Unit |
FREE_COB |
Boolean indicating if the contaminant COBis a free parameter of the PSF fitting model |
bool |
[-] |
COB_TS |
Centre of brightness time series of the contaminant |
CENTRE_OF_BRIGHTNESS Struct |
[-] |
Measurement status
Decimal Value |
Bit order |
Bit value |
Name |
Description |
|---|---|---|---|---|
0 |
0 |
00000000 00000000 |
VALID |
Nominal value (nothing to report) |
1 |
0 |
00000000 00000001 |
INVALID |
Invalid for any reasons |
2 |
1 |
00000000 00000010 |
MASK_CHANGED |
Mask changed |
4 |
2 |
00000000 00000100 |
ONG_LC_OUTLIERS |
Outliers detected on-ground over light-curves (both for light curves coming from on-board or light-curves coming from photometry on imagettes). Value considered as an outlier by the on-ground outlier detection algorithm over light-curves (ONG-OUTLCCOR-010) |
8 |
3 |
00000000 00001000 |
ONG_IMG_OUTLIERS |
Outliers detected on-ground over imagettes. Value considered as an outlier by the on-ground outlier detection algorithm over imagettes (ONG-OUTIMGCOR-010) One or several outliers detected in the imagettes from which the flux was extracted The detection of one or several outliers in the imagettes does not necessarily mean that the flux value extracted from the imagette is invalid |
16 |
4 |
00000000 00010000 |
RW_OFFLOADING |
Measurement acquired during a reaction wheel offloading |
32 |
5 |
00000000 00100000 |
ONB_LC_OUTLIERS |
Outlier(s) detected and removed on-board Relevant for 50s and 600s light-curves computed on-board. This flag does not necessarily mean that the flux value is invalid |
64 |
6 |
00000000 01000000 |
|
|
128 |
7 |
00000000 10000000 |
|
|
256 |
8 |
00000001 00000000 |
|
|
512 |
9 |
00000010 00000000 |
|
|
1024 |
10 |
00000100 00000000 |
|
|
2048 |
11 |
00001000 00000000 |
|
|
4096 |
12 |
00010000 00000000 |
|
|
8192 |
13 |
00100000 00000000 |
ERROR_BKG_MOD |
Computation error for bkg model |
16384 |
14 |
01000000 00000000 |
ERROR_LTDJIT_CORR |
Computation error for jitter and long term drif correction |
32768 |
15 |
10000000 00000000 |
COMPUTATION_ERROR |
Computation error value |
65536 |
16 |
00000001 00000000 00000000 |
ERROR_GAIN_VARIATION |
Computation error for gain variation |
131072 |
17 |
00000010 00000000 00000000 |
MERGING_NO_VALID |
Value not valid after merging |
Sky positions data product
Metadata
Sky position of a single target metadata
|
Description |
Type |
Unit |
WINDOW_ID |
Id of the window |
uint32 |
[-] |
PLATOID |
PLATO ID of the target |
uint64 |
[-] |
CAMERA_ID |
ID of the camera (1 to 26) |
uint8 |
[-] |
CCD_ID |
ID of the CCD
|
uint8 |
[-] |
SAMPLING |
Sampling of the background model time-series (typical value 25s) |
double |
second |
CONTAMINANTS_LIST |
List of instance of the CONTAMINANTPCOB structure. Each instance gives some information related to a given neighborhood star. Empty list if no contaminant stars. |
CONTAMINANTPCOB struct
(Nc) |
[-] |
COB_ORIGIN |
If relevant, bit array giving the origin of the COB used to derive this sky position |
uint8 |
[-] |
C_PROCESSING |
If relevant, bit array giving the processing undergone by the COB used to derive this sky position |
uint16 |
[-] |
I_PROCESSING |
Bit array giving the processing undergone by the imagette , relevant if this COB comes from aperture photometry on on-ground processed imagette |
uint16 |
[-] |
Data contain
Sky position of a single target for an exposure time
|
Description |
Type |
Unit |
BARYCENTRIC_TIME |
Timestamp (barycentric) when the sky position (or measurements to derive the sky position) has been acquired onboard. Expressed in Julian days since J2000_TIME_ORIGIN (i.e. ONBOARD_TIME=0). |
double |
Julian day |
ONBOARD_TIME |
Timestamp based on the on-board time when the sky position (or measurements to derive the sky position) has been acquired onboard. Expressed in Julian days since J2000_TIME_ORIGIN (i.e. ONBOARD_TIME=0). |
double |
second |
GCRS_RA |
RA sky position of the target in GCRS inferred from its measured CCD position |
double |
degrees |
GCRS_DEC |
DEC sky position of the target in GCRS inferred from its measured CCD position |
double |
degrees |
GCRS_DELTA_LONG |
Longitudinal displacement (tangential shift in the Right Ascension direction) in the GCRS frame. GCRS_DELTA_LONG = cos(Dec) * ( CCD_Ra - Sky_Ra), where:
|
double |
arcseconds |
GCRS_DELTA_LONG_ERR |
1-σ uncertainty associated with GCRS_DELTA_LONG |
double |
arcseconds |
GCRS_DELTA_LAT |
Latitudinal displacement (tangential shift in the Declination direction) in the GCRS frame. GCRS_DELTA_LAT = (CCD_Dec - Sky_Dec), where:
|
double |
arcseconds |
GCRS_DELTA_LAT_ERR |
1-σ uncertainty associated with GCRS_DELTA_LAT |
double |
arcseconds |
STATUS |
Status of the measurement (See STATUS Definition) |
uint32 |
[-] |
PSF data product
|
Description |
Type |
Unit |
PSF_ID |
PSF ID |
uint32 |
[-] |
PLATOID |
PLATO ID of target |
uint64 |
[-] |
RESOLUTION |
Resolution as a fraction of a pixel. Typical value: 128 (corresponding to a resolution of 1/128th of a pixel) |
uint32 |
[-] |
PSF |
PSF, result of inversion process |
double (M*M) |
[-] |
SIZE |
PSF size, i.e. physical number of pixels on a side of the PSF array |
uint8, typically 6 |
pixel |
B_SPLINE_RESOLUTION |
Resolution of the B-Spline representation of the PSF. Typical value: 20 corresponding to a resolution of 1/20th of pixel (in both the x and the y direction) |
uint32 |
[-] |
B_SPLINE_COEFFICIENTS |
B-Spline representation
of the PSF. The number of elements N in each direction depends
on B_SPLINE_RESOLUTION. |
double (N*N) |
[-] |
SPLINE_DEGREE |
Degree of splines (Typical value: 3 for cubic splines) |
uint8 |
[-] |
KNOT_TYPE |
Strategy to place the knots within the PSF. Values: SIMPLE (0) or DIERCKX (1). (Typical value: DIERCKX) |
uint8 |
[-] |
BARYCENTRIC_TIME |
Timestamp (barycentric) when the imagette do derive PSF has been acquired onboard. Expressed in Julian days since J2000_TIME_ORIGIN (i.e. ONBOARD_TIME=0). |
double |
Julian day |
ONBOARD_TIME |
Timestamp based on the on-board time when the imagette do derive PSF has been acquired onboard. Expressed in seconds since J2000_TIME_ORIGIN. |
double |
second |
CENTER |
x center and y center of the PSF within the grid (6*6) |
double (2) |
pixel |
COVARIANCE |
Covariance matrix of PSF enabling us to calculate the extent of the PSF in any direction. |
double (2*2) |
pixel² |
CAMERA_ID |
ID of the camera (1 to 26) |
uint8 |
[-] |
CCD_ID |
ID of the CCD
|
uint8 |
[-] |
CCD_POSITION |
CCD position of the target star using the CCD-RF CCD_POSITION_X = CCD_POSITION[0] CCD_POSITION_Y = CCD_POSITION[1] |
double (2) |
pixel |
FP_POSITION |
Position of the target star within the Focal Plane FP_POSITION_X = FP_POSITION[0] FP_POSITION_Y = FP_POSITION[1] |
double (2) |
mm |
NORM |
The norm of the PSF prior to normalisation. |
double |
e- |
REG_PARAMETER |
Non-dimensional version of the regularisation parameter used when carrying out the wPRLS inversion. Dividing this parameter by (NORM/SIZE2)2 produces the dimensional version of the regularisation parameter that is used in the inversion program.
|
double |
[-] |
FIT_ERROR |
Data fit error from the inversion process. This corresponds to the first term in the cost function for the wPRLS inversion. |
double |
[-] |
REG_ERROR |
Regularisation penalty. This measures the degree to which the inverted PSF is not regularised and corresponds to a normalised version of the second term in the cost function for the wPRLS inversion. |
double |
[-] |
STATUS |
Validation status of PSF (see PSF_STATUS) |
uint8 |
[-] |